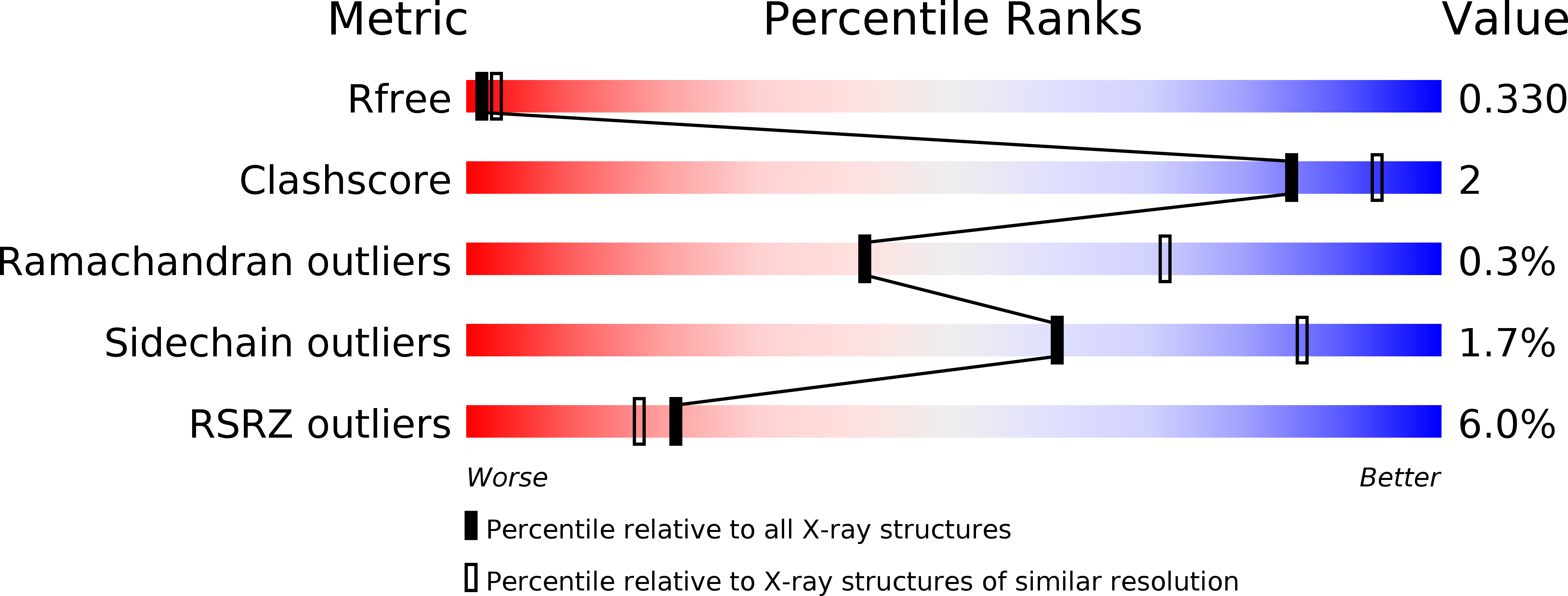
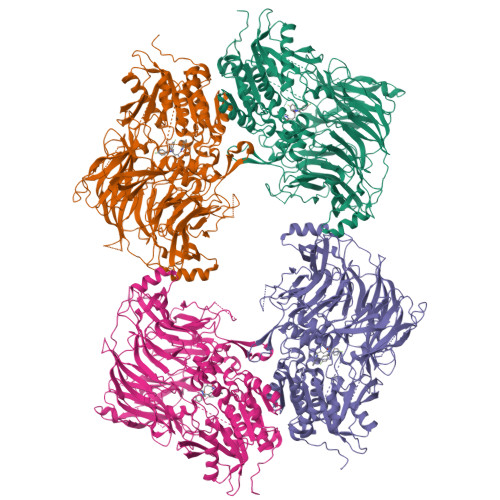
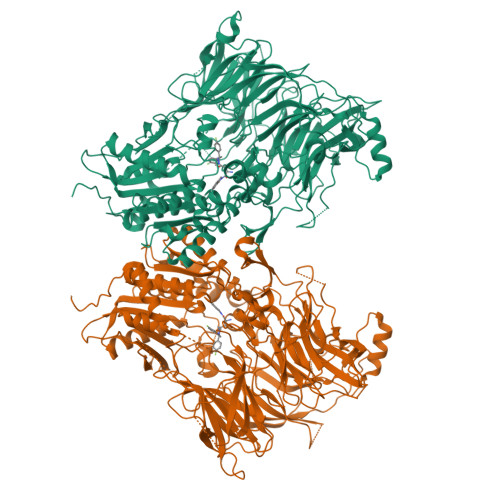
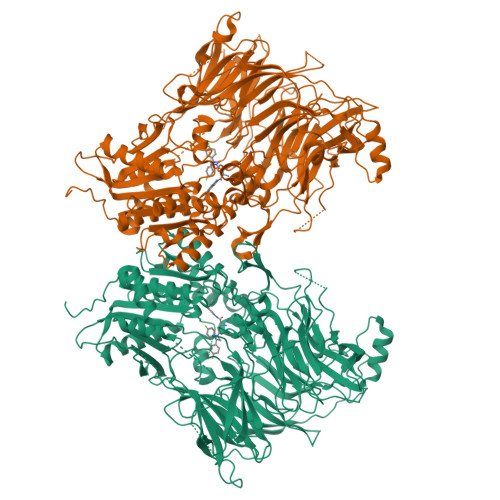
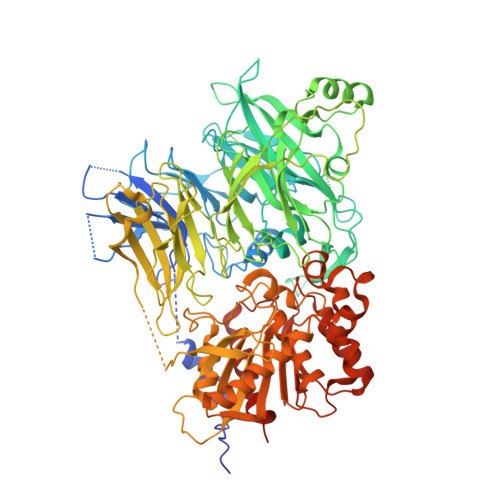
Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Ross, B., Krapp, S., Augustin, M., Kierfersauer, R., Arciniega, M., Geiss-Friedlander, R., Huber, R.(2018) Proc Natl Acad Sci U S A 115: E1437-E1445
- PubMed: 29382749
- DOI: https://doi.org/10.1073/pnas.1717565115
- Primary Citation of Related Structures:
6EOO, 6EOP, 6EOQ, 6EOR, 6EOS, 6EOT - PubMed Abstract:
Dipeptidyl peptidases 8 and 9 are intracellular N-terminal dipeptidyl peptidases (preferentially postproline) associated with pathophysiological roles in immune response and cancer biology. While the DPP family member DPP4 is extensively characterized in molecular terms as a validated therapeutic target of type II diabetes, experimental 3D structures and ligand-/substrate-binding modes of DPP8 and DPP9 have not been reported. In this study we describe crystal and molecular structures of human DPP8 (2.5 Å) and DPP9 (3.0 Å) unliganded and complexed with a noncanonical substrate and a small molecule inhibitor, respectively. Similar to DPP4, DPP8 and DPP9 molecules consist of one β-propeller and α/β hydrolase domain, forming a functional homodimer. However, they differ extensively in the ligand binding site structure. In intriguing contrast to DPP4, where liganded and unliganded forms are closely similar, ligand binding to DPP8/9 induces an extensive rearrangement at the active site through a disorder-order transition of a 26-residue loop segment, which partially folds into an α-helix (R-helix), including R160/133, a key residue for substrate binding. As vestiges of this helix are also seen in one of the copies of the unliganded form, conformational selection may contributes to ligand binding. Molecular dynamics simulations support increased flexibility of the R-helix in the unliganded state. Consistently, enzyme kinetics assays reveal a cooperative allosteric mechanism. DPP8 and DPP9 are closely similar and display few opportunities for targeted ligand design. However, extensive differences from DPP4 provide multiple cues for specific inhibitor design and development of the DPP family members as therapeutic targets or antitargets.
Organizational Affiliation:
Max Planck Institut für Biochemie, D-82152 Martinsried, Germany; ross@biochem.mpg.de huber@biochem.mpg.de.